I remember the astonishing diversity of life in a drop of lake water I first viewed under a microscope sixty years ago. Now we use cryogenic electron microscopes, DNA sequencers, protein splicers, and supercomputers to create computer simulations of life down to viruses. In Yinbin, China, two exceedingly bright bioinformatics researchers have created a 3-D virtual model of COVID–19 and have been testing virtual medications against it. In the process, they identified a binding site on human red blood cells for a COVID-19, which changes red cell architecture and function. The genome for COVID-19 was published January 20, 2020. The Yinbin model was posted 60 days later.
Ten years ago, such a model would take years.
During the same period, Critical Care physicians at Beth Israel – Deaconess Medical Center noted that patients’ intravenous lines were clotting frequently and quickly. They obtained emergency regulatory approvals to administer blood thinners to COVID-19 patients about the same time the Yinbin model was published, demonstrating abnormal red cell function.
Critical care doctors in New York noticed that they had patients who could walk and talk with phenomenally low oxygens. They stuck with using noninvasive techniques to raise oxygen far longer than previously. A fair proportion of patients weathered their COVID-19 storm without ventilator use. The Yinbin model had also indicated that when COVID-19 attacked a red blood cell, it altered its ability to carry oxygen.
This is science on the march. Knowledge and intuition shared without boundaries. It’s not a step-by-step process; it is hundreds of researchers and physicians, simultaneously contributing pieces to a puzzle that will clarify the functions of COVID-19 so we can stop the pandemic carnage.
The Yinbin model is also used to screen drug candidates. It can scan millions of possible binding sites per second to see if there is a three-dimensional fit between drug and coronavirus. Both hydroxychloroquine and favipiravir (Avgen), a Japanese antiviral like Tamiflu, show an affinity to the red cell where the coronavirus protein attaches, and could competitively block that attachment. There are ten more drugs under consideration which await clinical evaluation for conclusions.
“Trace Together” – Contact Tracing at Light Speed
In the 1918 Spanish Flu pandemic, New York had had the best management of US cities. It had lower death rates than Philadelphia and San Francisco. It did not have a second wave like St. Louis. New York had a massive public health infrastructure because it was the point of entry of major immigrant communities. New York had the personnel to perform contact tracing and quarantine. Singapore has distributed a contact tracing cellphone app (Trace Together). It uses Bluetooth to log every nearby (6ft) phone you’ve been close to for over 30 minutes. If one is diagnosed with COVID-19, their contacts are notified to report for testing. Google and Apple have announced a joint task force to create a similar app for United States cellphones. If this process is successful, contact tracing and quarantine can cripple this pandemic.
A Vaccine Can Defeat COVID-19
The logical target for a vaccine is to block the protein that attaches COVID-19 to our respiratory cells. Those red “spikes” (above) bind to the ACE protein on our respiratory cells. Without a cell to supply an environment, the virus cannot spread. Jason McLellan, associate professor at UT Austin, and his colleague had spent many years studying other coronaviruses, including SARS-CoV and MERS-CoV. They had already developed methods for analyzing coronavirus spike proteins that could turn them into candidates for vaccines. “We knew exactly what mutations to put into this, because we’ve already shown these mutations work for other coronaviruses,” said McLellan.
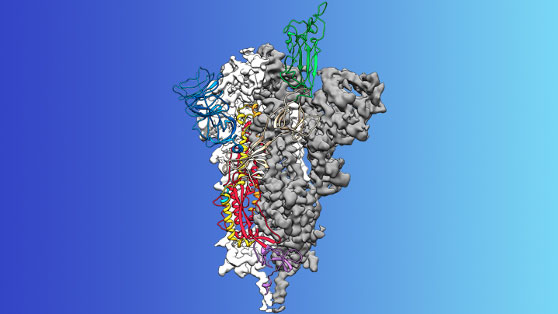
Two weeks after receiving a genome sequence of the virus from Chinese researchers, his team designed and produced samples of their stabilized spike protein. It took approximately 12 more days to reconstruct the 3D atomic scale map of the spike protein, which you can see above (credit to UT) and submit a manuscript. The small area of green at the top of the map is the attachment spike.
We are developing over 200 treatments and vaccines. University of Pittsburgh, with knowledge of the spike protein composition, has manufactured a skin patch permeated with spike protein, which stimulates manufacture of antibodies against the virus. The patch would have far fewer side effects than a classic vaccine injection and easier to distribute. It would also have a level of specificity greater than almost all previous vaccines.
The US Military, in conjunction with a partnership of the major pharmaceutical vaccine firms, is testing a vaccine which manufacture antibodies against the virus (as opposed to stimulating the patient’s body to do the job). The stimulus lasts eight weeks. This would be a cross between treatment and vaccination if the antibody is long-lived.
In 1952, Jonas Salk grew poliovirus into African green monkeys, and then took the virus and killed it with formaldehyde. He then tested in two groups of school children; he also took hypodermics home, boiled them on the kitchen stove, and injected his wife, his children, and himself. In 1954. When asked to which drug companies (or the March of Dimes) he would grant the patent, he replied that “one could not patent the sun,” and gave the patent to the world. This is the level of commitment of the scientists, the clinicians, the healthcare workers, and first responders in this fight.
References:
Illustrations: courtesy NIH
Next Article: Finding Coronavirus where It Hides